Research
Our Philosophy: Cutting Edge Proteomics and Molecular Biology - an Ideal Partnership for a Better Understanding of the Lysosome
The Winter Lab is investigating molecular mechanisms related to the function of lysosomes. We are using state of the art proteomics, molecular biology and biochemistry approaches to characterize proteins located at the lysosome and to investigate how they change in response to different stimuli. Our aim is not only to identify changes on these proteins using mass spectrometry based proteomics approaches, but also to characterize the role these changes play for the function of the lysosome.
For further information please visit:
The Lysosome
The lysosome is the degradative compartment of the cell. Present in almost any living cell, it is degrading macromolecules and organelles and organizing the recycling of cellular components. Furthermore, it is involved in host defense mechanisms, receptor mediated endocytosis and antigen presentation to name just a few of the plethora of mechanisms it is involved in. Currently, around 150 lysosomal proteins are confirmed and the list is constantly growing.
The lysosomal lumen – a place for degrading (almost) everything
The inside of the lysosome, its lumen, is home to a variety of hydrolases which catalyze various degradation reactions. Almost every molecule which is important to keep a cell functional can be broken down here to its building blocks. Like a molecular shredder, it can take on everything ranging from single molecules to whole organelles (for example mitochondria). These large molecules are then degraded to their building blocks which are lipids, sugars, proteins and nucleotides.
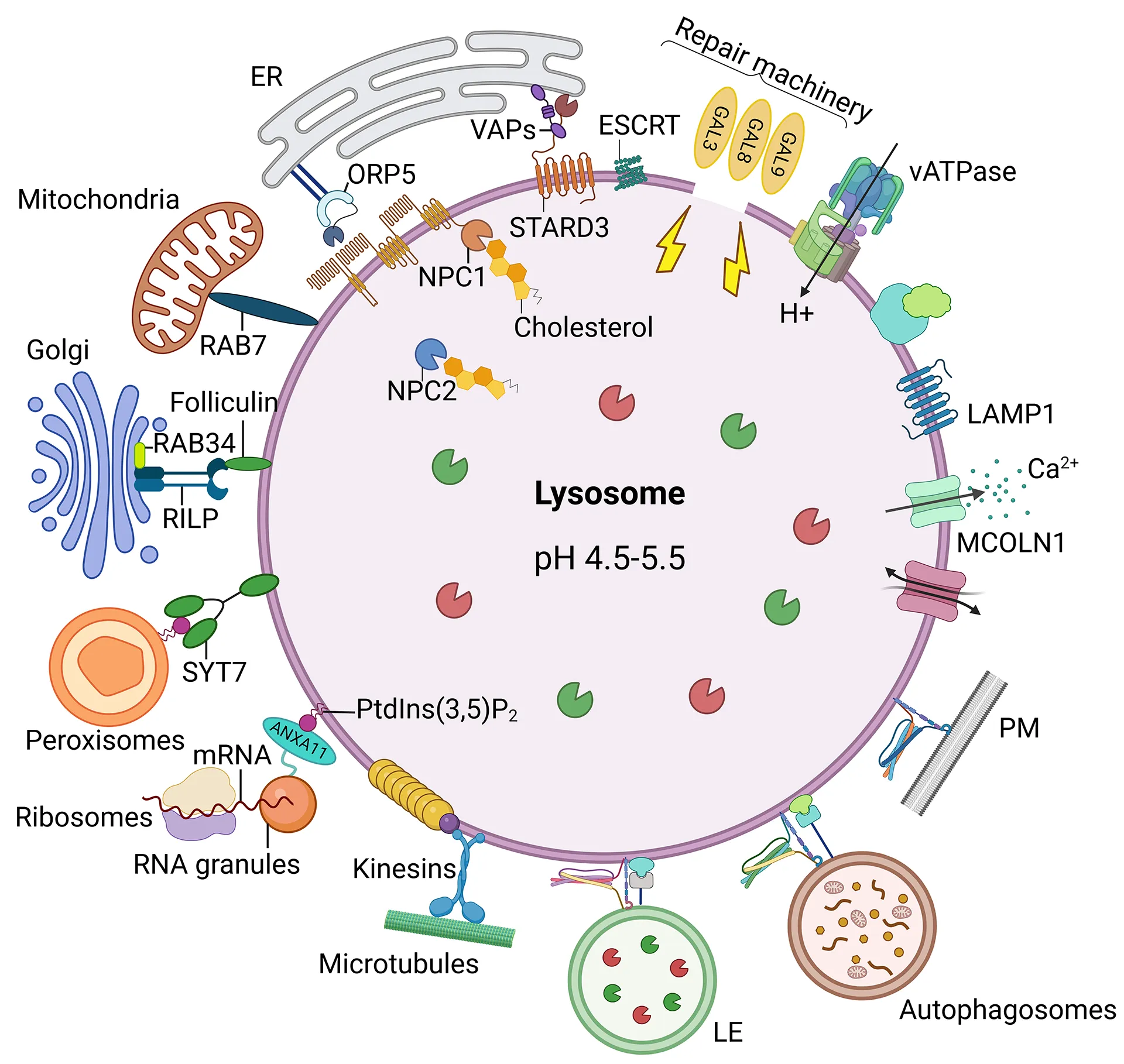
The lysosomal membrane – the gate to the lysosome and harbor for signaling molecules
The molecules located in the lysosomal membrane take on various functions being important for its structure, acidification, localization in the cell, import of molecules to be degraded, fusion with other organelles and transport of the degradation products to the cytosol. Various proteins also serve as docking station for others and there is an emerging picture of membrane proteins interacting with each other. Despite decades of research, for many lysosomal membrane proteins the function is still unclear and constantly novel functions are discovered making the field of lysosomal research an exciting and vibrant area.
Lysosomal Storage Disorders – A unique class of severe diseases
Malfunctions of each of the lysosomal hydrolases result in severe inherited diseases termed lysosomal storage disorders (LSDs). The common hallmark of these diseases is the accumulation of not degraded material in the lysosome which impairs its function. Currently, about 50 LSDs are known with a cumulative incidence of ~ 1 in 5.000 new-borns. A prominent subgroup of LSDs originates from defects in sphingolipid degrading enzymes resulting in sphingolipid storage diseases such as Gaucher disease, Fabry disease or metachromatic leukodystrophy (MLD). Despite many years of research, the effects of these disorders on the cellular level are still poorly understood.
Our approach to understand the lysosome and related disorders
To gain more insight in the processes involved leading to malfunction of the lysosome, we are investigating changes in whole cell or lysosomal protein composition as a consequence of increasing lipid storage. For this purpose, we analyze animal and cell culture models using state of the art liquid chromatography tandem mass spectrometry methods. We also establish novel methods, if needed, and apply them to model systems performing mainly quantitative analysis of proteins and their posttranslational modifications.
Our toolbox
Stable isotope based labeling strategies like SILAC (stable isotope labeling of amino acids in cell culture), TMT (tandem mass tags) or dimethyl labeling play a prominent role in our research. These methods allow us to simultaneously identify and quantify thousands of proteins and their posttranslational modifications in one mass spectrometry experiment. In order to deal with the high complexity of the samples, we perform enrichment and fractionation of peptides and proteins using a variety of methods. Once we identify interesting candidate proteins in these experiments, we conduct follow up studies using a variety of approaches. We confirm protein expression levels using western blots, qPCR or enzyme assays and once we are convinced that we found something interesting, we investigate cells using immunohistochemistry and perform knockdown, overexpression or mutation studies, just to name the most important.


