Research
The Proteostasis Network
In order to fulfil their biological function proteins must fold into their native three-dimensional structures. Aberrant folding or unfolding compromises the functionality of the affected protein and is accompanied by a great risk of disrupting other proteins by undergoing non-specific protein-protein interactions. Especially metastable proteins with disordered regions (for example up to 30% of the mammalian proteome) are prone to undergo unwanted interactions and form toxic protein aggregates, which are associated with neurodegenerative diseases. Thus, cells must ensure either proper protein folding, or if this fails, efficient degradation of non-native proteins. The factors controlling these processes are collectively termed the protein homeostasis (proteostasis) network. At the center of the proteostasis network are molecular chaperones, which assist in protein folding and prevent aggregation, refold stress-denatured proteins and cooperate with the ubiquitin proteasome system (UPS) and the autophagy pathway in the degradation of terminally misfolded proteins.
Cellular Compartmentalization of Protein Quality Control
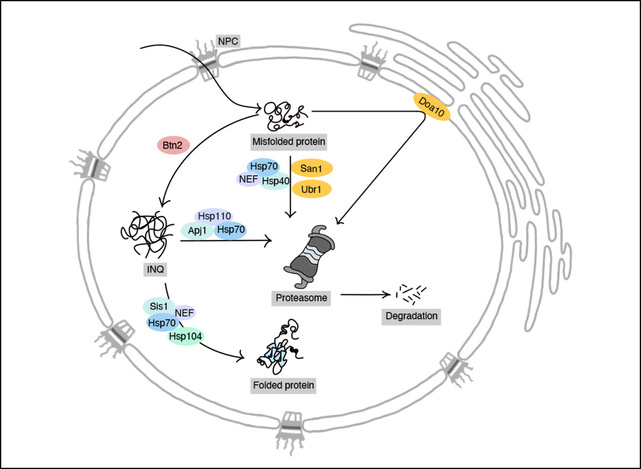
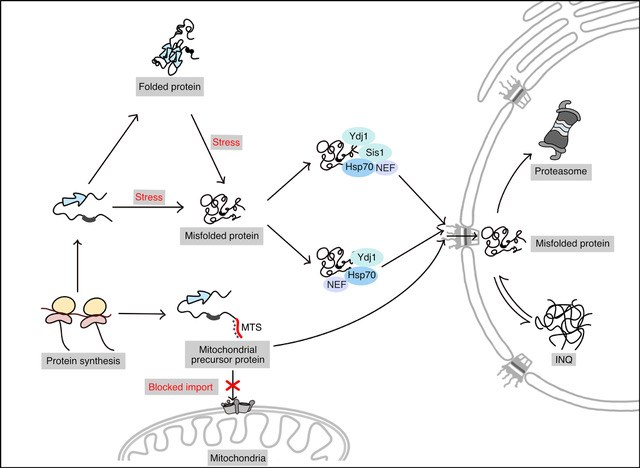
We are in particular interested in the interplay of different subcellular compartments in protein quality control. Most compartment have a dedicated set of factors for removing or repairing defective proteins. For instance, the cytosol contains specific pathways for the turnover of misfolded proteins depending on their aggregation state. Soluble proteins are typically turned over by the 26S proteasome, while larger protein aggregates, not accessible to the proteasome, are targeted for degradation by autophagy. Using the yeast S. cerevisiae as a model system we had previously determined the criteria for pathway choice between those two pathways acting in the cytosol (Lu et al., 2017). The autophagic machinery is absent from the nucleus, which thus requires an alternative mechanism for aggregate clearance. We identified the Hsp40 chaperone, Apj1, as a specific nuclear chaperone required for resolving aggregated proteins and their subsequent targeting for degradation (den Brave et al., 2020). An intriguing discovery during our studies on nuclear protein quality control was the presence of many proteins of other subcellular compartments in nuclear protein aggregates, including mitochondrial proteins. This might suggest that different cellular compartments interact to buffer defects in protein quality control. Our goal is to understand the underlying mechanisms and physiological benefits of different compartments interacting in proteostasis.
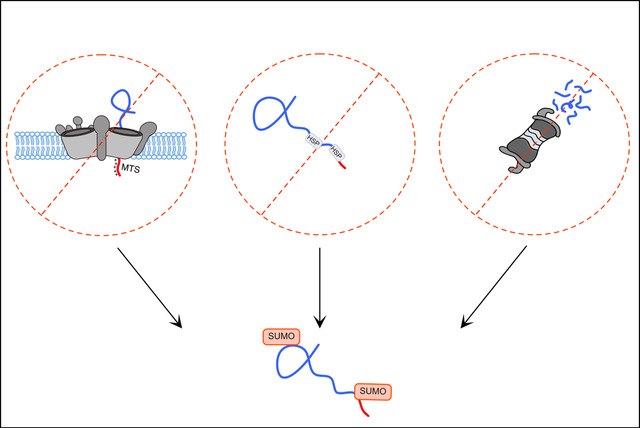
Ubiquitin-like modifications in Proteostasis
Efficient protein quality control requires mechanism of substrate recognition and tagging for subsequent processing by different pathways. A central question of our research is how such sorting is mediated by ubiquitin-like proteins. For example, ubiquitin itself is well-known for its role in targeting proteins to the main proteolytic pathways, the ubiquitin proteasome system and the autophagic machinery. Autophagy in addition largely depends on another ubiquitin-like protein called Atg8/LC3. We mainly focus on the role of the ubiquitin-like protein SUMO in protein quality control. Modification of proteins with SUMO is strongly induced by proteotoxic stress, i.e. conditions causing proteins to misfold. Moreover, the proteins aggregating in the most common age-related neurodegenerative diseases have been found to be modified by the small ubiquitin-like modifier SUMO, including, Alzheimer’s, Parkinson’s and Huntington’s disease. Despite these well-established links to protein quality control, the role of the SUMO pathway in proteostasis is only poorly understood. We aim to understand the role of SUMOylation in response to proteotoxic stress and in the context of protein aggregation. Moreover, we study the role of SUMOylation on different cellular compartments. In fact, the SUMO-pathway and its substrates are predominantly localized inside the nucleus, but also cytoplasmic targets have been described. We have identified mitochondrial proteins as a novel class of SUMOylated substrates linked to protein quality control (Paasch et al., 2018). We aim at understanding the role of SUMOylation on mitochondrial proteins and the general function of the SUMO pathway within the proteostasis network.